В журнале Journal of the American Chemical Society (JACS), (IF 16,383) опубликована статья с участием сотрудников Института: C.C. Овчеренко (мнс, ЛМР), к.х.н. А.В. Шернюкова (снс, ЛМР), Д.М. Насонова (лаборант, ЛМР) и д.ф.-м.н., проф. Е.Г. Багрянской (Директор НИОХ)
Dynamics of 8-Oxoguanine in DNA: Decisive Effects of Base Pairing and Nucleotide Context
Sergey S. Ovcherenko, Andrey V. Shernyukov, Dmitry M. Nasonov, Anton V. Endutkin, Dmitry O. Zharkov*, and Elena G. Bagryanskaya*
J. Am. Chem. Soc. 2023, 145, 10, 5613-5617
Publication Date:March 2, 2023
doi: 10.1021/jacs.2c11230
Abstract
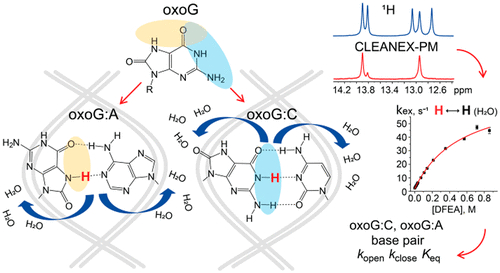
8-Oxo-7,8-dihydroguanine (oxoG), an abundant DNA lesion, can mispair with adenine and induce mutations. To prevent this, cells possess DNA repair glycosylases that excise either oxoG from oxoG:C pairs (bacterial Fpg, human OGG1) or A from oxoG:A mispairs (bacterial MutY, human MUTYH). Early lesion recognition steps remain murky and may include enforced base pair opening or capture of a spontaneously opened pair. We adapted the CLEANEX-PM NMR protocol to detect DNA imino proton exchange and analyzed the dynamics of oxoG:C, oxoG:A, and their undamaged counterparts in nucleotide contexts with different stacking energy. Even in a poorly stacking context, the oxoG:C pair did not open easier than G:C, arguing against extrahelical base capture by Fpg/OGG1. On the contrary, oxoG opposite A significantly populated the extrahelical state, which may assist recognition by MutY/MUTYH.
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jacs.2c11230.
-
Additional experimental details, description of proton exchange formalism and NMR experiments, supplementary discussion, as well as supplementary Tables and Figures including additional experimental results (PDF)
Альметрики:
Метрики PlumX теперь доступны в Scopus: узнайте, как другие ученые используют ваши исследования

